News
Two nf-core events supported by BioDATEN
- Published on Friday, 24 September 2021
BioDATEN supports two nf-core events helt and organized by QBiC:
- nf-core bytesize talks: talks organized by the nf-core community on how to write Nextflow pipelines with the help of the nf-core tools. This series will also include information on currently available analysis pipelines as part of the nf-core project and how to use them. The talks from the previous series are available in our YouTube channel: https://www.youtube.com/watch?v=ZfxOFYXmiNw&list=PL3xpfTVZLcNiSvvPWORbO32S1WDJqKp1e
- nf-core hackathon: a developer event to contribute to the nf-core Nextflow pipelines. This year we will focus on converting existing pipelines to Nextflow DSL2
(GG)
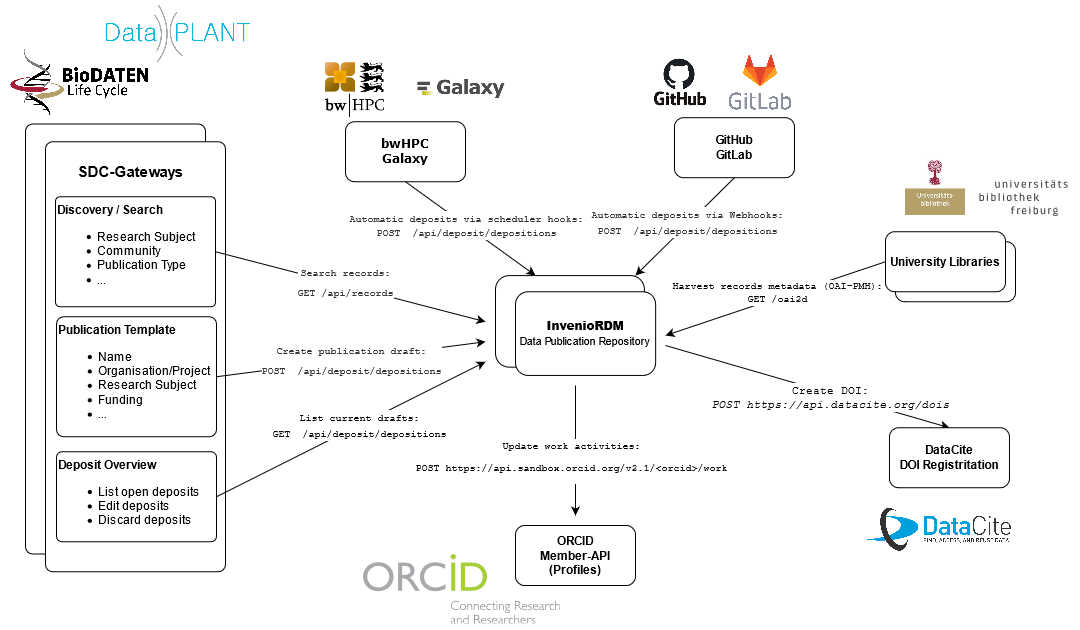
Science Data Center infrastructure: BioDATEN implements InvenioRDM for data publication
- Published on Wednesday, 07 July 2021
Jonathan Bauer from the University of Freiburg gave a presentation about the joined effort of establishing a publication repository for the BioDATEN community. The Invenio framework for data publication is arriving at its first release status. BioDATEN is getting into a partnership with the developers to push forward the productive version. Authentication is handled through KeyCloak and an object storage backend is offered through bwSFS. Currently a standalone instance is deployed but a Kubernetes-based orchestration solution is currently evaluated to cope with future usage load. Login is possible via ELIXIR ID authenticating against the identity provider of their home institutions. KeyCloak facilitates a role-based access control. A strategy for reproducible deployment, resilient operation and a disaster recovery is currently developed. Records in the repository can be individually controlled for data and metadata. An embargo or holding period can be applied to a record which is not publicly available until the embargo has ended. Special viewing and edit links can be generated and shared. A range of persistent identifiers can be applied, primarily support for the DataCite DOIs. Its possible to specify existing DOIs or mint new ones. Additionally, further identifiers can be specified like ARK, arXiv or handle. The integration with 3rd-party services is exposed through a REST API. This enables significant integration potential with other services like auto-updating of profiles with new publications, integration with the Science Gateway of BioDATEN for discovering or the publication of templates or for GitLab instances webhooks could be used to publish code releases.
(DvS, JK)
KeyCloak as an authentication proxy for BioDATEN
- Published on Wednesday, 07 July 2021
Identity and access management is a core component for BioDATEN as for all research data management infrastructures. Jonathan Bauer from the University of Freiburg just summarized the state of developments within BioDATEN for services like Invenio or Hubzero for a presentation for the SDC infrastructure working group. The main challenge is the interplay of services currently developed for the BioDATEN community. A contemporary user experience relies on a single-sign on concept, enabling users to access all BioDATEN services with just one set of credentials. BioDATEN relies on the ELIXIR AAI as pan-european infrastructure providing EXLIXIR IDs for its users. KeyCloak is an open source framework for identity and access management with single-sign on functionality. It supports many common identity providers and protocols like e.g. Active Directory, LDAP, SAML or OpenID-Connect, allowing identity brokering and linking of multiple identities. The framework is capable of handling different forms of authorisation and is very customizable for this purpose. The aim for BioDATEN is the establishment of trust delegation among BioDATEN services via KeyCloak. Services shall interact with each other on the users behalf e.g. allowing the submission of a bioinformatic analysis workflow from the BioDATEN portal which uses a dataset as input which is stored within the Invenio publication repository.
(DvS, JK)
